🌋 EM27 Retrieval Pipeline: Automated EM27/SUN Data Processing
We retrieve a lot of EM27/SUN data, produced mainly by MUCCnet (Dietrich et al., 2021), and have used this pipeline since end of 2021.
This codebase provides an automated data pipeline for Proffast 1 and 2.X. Under the hood, it uses the Proffast Pylot to interact with Proffast 2 and an in-house connector to interact with Proffast 1. Whenever using this pipeline for Proffast retrievals, please make sure to also cite Proffast and the Proffast Pylot (for Proffast 2.X retrievals).
📚 Read the documentation at em27-retrieval-pipeline.netlify.app.
💾 Get the source code at github.com/tum-esm/em27-retrieval-pipeline.
🐝 Report Issues or discuss enhancements using issues on GitHub.
Related Projects:
⚙️ Many of our projects (including this pipeline) use functionality from the tum-esm-utils package.
🤖 Our EM27/SUN systems are running autonomously with the help of Pyra.
EM27 Retrieval Pipeline vs. PROFFASTpylot
This pipeline does not intend to replace the Proffast Pylot by any means. It uses most of the Pylot’s functionality and provides an automation layer on top of that. You can use Proffast directly for full flexibility, the Pylot for some flexibility, or this pipeline, which is more opinionated than the Pylot but highly automated. Furthermore, this pipeline will continue to include support for the different retrieval algorithms.
We decided to include a copy of the Python codebase of the Proffast Pylot inside this repository so we can have less complexity due to Git Submodules or on-demand downloads. Since the Proffast Pylot only supports Proffast 2.X, we have added our own wrapper for Proffast 1 in this pipeline.
What does this pipeline do?
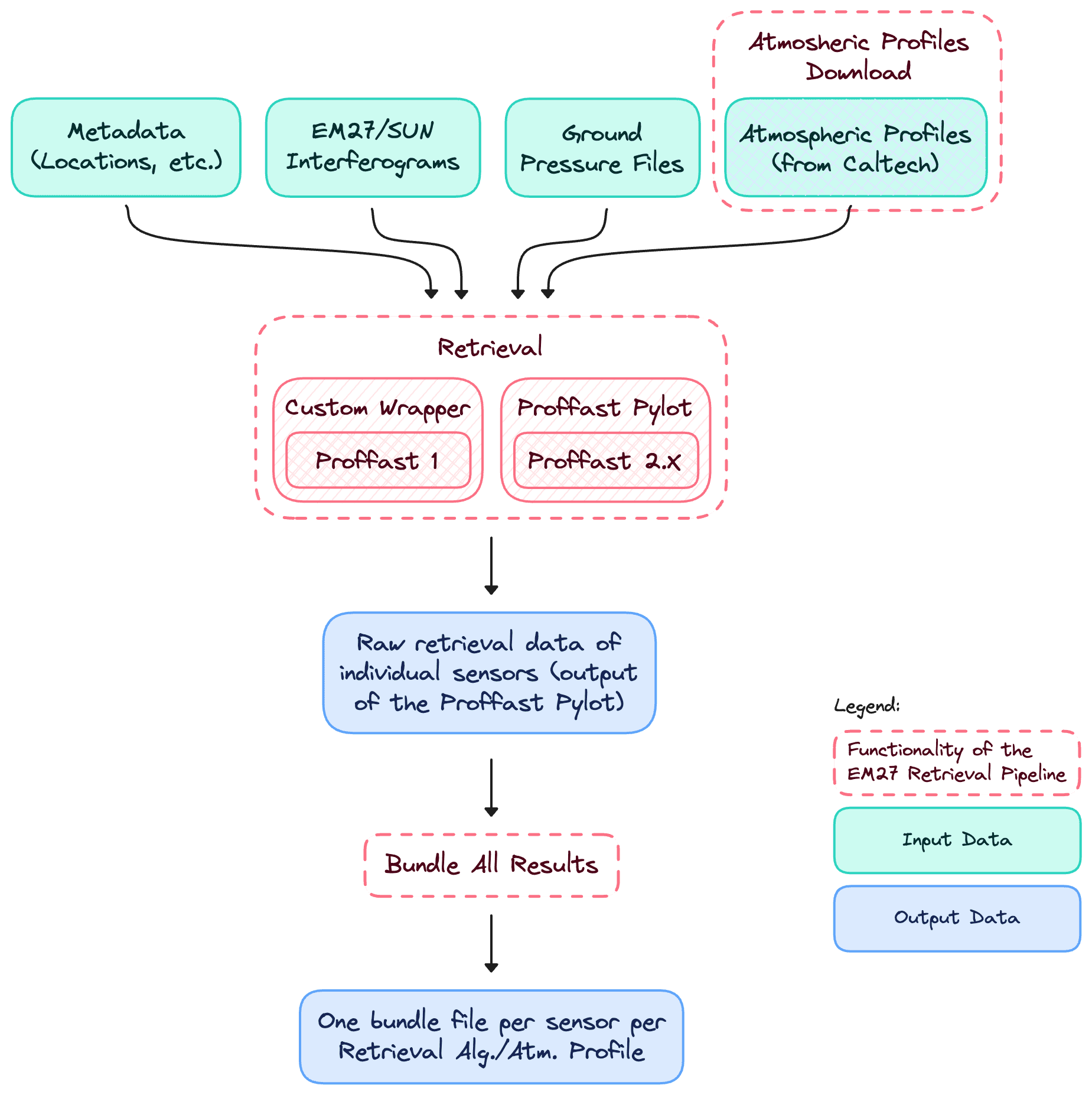
The pipeline consists of three building blocks that are required to retrieve EM27 data:
| Task | Script Entrypoint aaaaaaaaaaaaa |
|---|---|
| 1. Downloading vertical profiles from the Caltech FTP server | python cli.py profiles run |
| 2. Running the retrieval to generate the averaged column concentrations | python cli.py retrieval start |
| 3. Bundle all retrieval results into one file per sensor/retrieval alg./atm. profile | python cli.py bundle run |
The data flow from input to merged outputs:
The pipeline offers:
- Easy configuration of using a validated
config.json(and metadata files): By “validated”, we mean that before the processing starts, the config files content will be parsed and validated against a JSON schema. This way, you can be sure that the pipeline will not fail due to a misconfiguration, and you will immediately get precise error messages. - Opinionated management of station metadata: We manage our EM27 metadata using JSON files instead of database tables, which has several benefits mentioned in the metadata repository
- Filtering of interferogram files that Proffast cannot process: When some interferograms are corrupted, Proffast will fail during preprocessing for whole days of data even when only a few out of thousands of interferograms are bad. The pipeline filters out these interferograms and only passes the valid ones to Proffast. A standalone version of this filtering functionality is included in our utility library
- Parallelization of the Proffast Pylot execution: The Pylot already provides parallelization. However, we decided to isolate the approach of the Pylot more and run the retrieval execution for each station and date individually inside a containerized environment. This way, errors during the retrieval don’t affect separate days/stations, and we have separated outputs and logs.
- Fully automated interface to obtain Ginput Data: The atmospheric profiles downloader of this pipeline automates the request for GGG2014 and GGG2020 data (supporting standard sites) from
ftp://ccycle.gps.caltech.edu. The manual instructions can be found here. - Comprehensive logs and output data management: It will store failed and succeeded containers. The output is the same as with the Pylot but also contains all config files the pipeline used to run this container and logs generated by the container.
- Bundling of retrieval results: The raw retrieval outputs will be distributed over hundres or thousands of folders and files. The bundling script of this pipeline will merge these outputs into one file per station, retrieval algorithm, and atmospheric profile. This way, you can easily access the data for further processing. (Read about it here).
- Documentation and complete API reference: hosted at em27-retrieval-pipeline.netlify.app
Getting Started
To fully understand the pipeline, you should read the following sections:
- The configuration section explains how the pipeline uses a
config.jsonfile to read the parameters of your environment. - The directories section explains how the input directories should be structured and how the pipeline structures its outputs.
- The metadata section explains what the metadata the pipeline requires is structured and how to connect it to the pipeline.
- The usage section explains how to run the pipeline.
You can look into the API Reference pages for an exact schema definition and explanation of every parameter. You can also use this documentation’s search function (top right) to find what you are looking for.
If you have issues or feature requests, please open an issue on GitHub or ask Moritz Makowski (moritz.makowski@tum.de).




