Usage
Setting up the Pipeline
Get the code
- Clone the repository or download the source code of a specific release.
- Create a virtual environment using
python3.11 -m venv .venv - Always activate the virtual environment using
source .venv/bin/activate - Install the dependencies using
pip install ".[dev]"orpdm sync --group dev
Configure it on your system
- Install System Dependencies: e.g.
sudo apt install unzip gfortran - Use the
config/config.template.jsonto create aconfig.jsonfile. - Set up the metadata locally or remotely (see metadata guide)
Run the tests to ensure everything is working
- Test the pipeline using
pytest -m "integration or quick or ci" --verbose --exitfirst tests/ - Test the actual retrieval using
pytest -m "complete" --verbose --exitfirst tests/
Celebrate if the tests pass
- Run
curl parrot.live
Use the CLI to run the pipeline
The following sections describe how to trigger the different pipeline processes using the CLI. Whenever calling the CLI, it will validate the integrity of your local config.json file and output a message if your system is not correctly configured.

Downloading Atmospheric Profiles
This pipeline automated the way of obtaining atmospheric profiles described in https://tccon-wiki.caltech.edu/Main/ObtainingGinputData. Add the email address to access the FTP server to config.profiles.server.email and configure the scope you want to download in config.profiles.scope.
Run the following command to download all the profiles:
python cli.py profiles runThis script will use the metadata and the configured local profiles directory to determine which profiles to request. Every time you call this script, it will request the profiles it has not already requested and check for the results of ongoing requests.
This process ensures that only config.profiles.server.max_parallel_requests are
running simultaneously. It only requests the same profiles again if they have not been generated within 24 hours. The script can download partial query results (e.g. if only days 1 to 5 of a 7-day request could be fulfilled).
You can use config.profiles.GGG2020_standard_sites to configure a list of standard sites you want to download. The script will never request profiles for these standard sites but only download the pre-generated data.
Run the following to request the current queue status of your account:
python cli.py profiles request-ginput-statusRunning Retrievals
Use the following commands to start the retrievals in a background process.
python cli.py retrieval startYou can limit the number of cores used by the retrieval process using config.retrievals.general.max_process_count.
Using the following commands, you can check whether the retrievals are still running and open a dashboard to monitor the progress.
python cli.py retrieval is-runningpython cli.py retrieval watch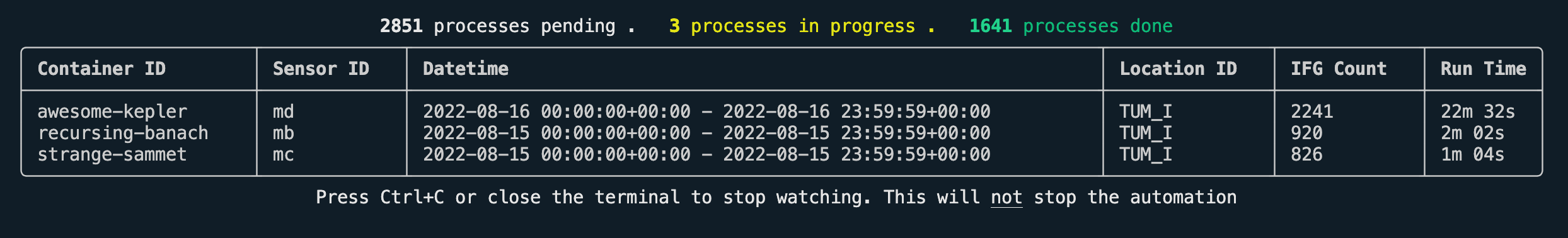
Terminate the ongoing retrievals using the following command:
python cli.py retrieval stopBundling All Retrieval Outputs
Bundle all the retrieval outputs using the following command:
python cli.py bundle runGenerating GEOMS compliant HDF5 files
The Generic Earth Observation Metadata Standard (GEOMS) is a standard for exchanging ground-based total-column concentration data. It uses the HDF5 file format and enforces a specific structure for the data (see guidelines from EVDC).
This pipeline can generate GEOMS compliant HDF5 files for the entire retrieval output. For that it requires the files calibration_factors.json
and geoms_metadata.json to be present in the configuration directory. You can use the example files and the API reference
to create your own files. You need to add a section geoms to the general config.json file defining
the scope for which you want to generate GEOMS files.
As always, the pipeline will tell you if any of your configuration files are invalid. Create the GEOMS files using the following command:
python cli.py geoms runThe logs will report, which files have been generated:
Config is validLoading configurationLoading calibration factorsLoading geoms metadataProcessing proffast-2.4/GGG2020Processing sensor id "ma"Sensor ma: found 1105 results in totalSensor ma: found 173 results within the time range ma/20240501: Generated .../proffast-2.4/GGG2020/ma/successful/20240501/groundbased_ftir.coccon_tum.esm061_munich.tum_20240501t114016z_20240501t171933z_001.h5 ma/20240502: Generated .../proffast-2.4/GGG2020/ma/successful/20240502/groundbased_ftir.coccon_tum.esm061_munich.tum_20240502t070418z_20240502t164544z_001.h5 ma/20240504: Generated .../proffast-2.4/GGG2020/ma/successful/20240504/groundbased_ftir.coccon_tum.esm061_munich.tum_20240504t053204z_20240504t171926z_001.h5 ma/20240505: Generated .../proffast-2.4/GGG2020/ma/successful/20240505/groundbased_ftir.coccon_tum.esm061_munich.tum_20240505t051558z_20240505t172421z_001.h5 ma/20240506: Generated .../proffast-2.4/GGG2020/ma/successful/20240506/groundbased_ftir.coccon_tum.esm061_munich.tum_20240506t052157z_20240506t125438z_001.h5 ma/20240507: Not enough data (less than 11 datapoints) ma/20240509: Generated .../proffast-2.4/GGG2020/ma/successful/20240509/groundbased_ftir.coccon_tum.esm061_munich.tum_20240509t072631z_20240509t152931z_001.h5 ma/20240510: Generated .../proffast-2.4/GGG2020/ma/successful/20240510/groundbased_ftir.coccon_tum.esm061_munich.tum_20240510t061155z_20240510t083149z_001.h5ma/20240511: 5%|██████ | 8/173 [00:19<03:07, 1.20s/it]You can verify the integrity of these HDF5 files using the AVDC’s Quality Assurance Tool or NILU’s GEOMS File Format Checker.
Generate a Data Report
You can generate a report about the data on your system using the following command:
python cli.py data-reportThis script will produce one CSV file per sensor ID in the directory data/reports/:
from_datetime,to_datetime,location_id,interferograms,ground_pressure,ggg2014_profiles,ggg2014_proffast_10_outputs,ggg2014_proffast_22_outputs,ggg2014_proffast_23_outputs,ggg2020_profiles,ggg2020_proffast_22_outputs,ggg2020_proffast_23_outputs2023-09-07T00:00:00+0000,2023-09-07T23:59:59+0000, TUM_I, 2224, 1440,✅,-,✅,✅,✅,-,✅2023-09-08T00:00:00+0000,2023-09-08T23:59:59+0000, TUM_I, 2178, 1440,✅,-,✅,✅,✅,-,✅2023-09-09T00:00:00+0000,2023-09-09T23:59:59+0000, TUM_I, 1966, 1440,✅,-,✅,✅,✅,-,✅2023-09-10T00:00:00+0000,2023-09-10T23:59:59+0000, TUM_I, 2034, 1440,✅,-,✅,✅,✅,-,✅2023-09-11T00:00:00+0000,2023-09-11T23:59:59+0000, TUM_I, 2122, 1440,✅,-,✅,✅,✅,-,✅2023-09-12T00:00:00+0000,2023-09-12T23:59:59+0000, TUM_I, 1972, 1440,✅,-,✅,✅,✅,-,✅2023-09-13T00:00:00+0000,2023-09-13T23:59:59+0000, TUM_I, 216, 1439,✅,-,✅,✅,✅,-,✅2023-09-14T00:00:00+0000,2023-09-14T23:59:59+0000, TUM_I, 762, 1440,✅,-,✅,✅,✅,-,✅2023-09-15T00:00:00+0000,2023-09-15T23:59:59+0000, TUM_I, 1507, 1440,✅,-,✅,✅,✅,-,✅2023-09-16T00:00:00+0000,2023-09-16T23:59:59+0000, TUM_I, 2232, 1440,✅,-,✅,✅,✅,-,✅2023-09-17T00:00:00+0000,2023-09-17T23:59:59+0000, TUM_I, 1599, 1440,✅,-,✅,✅,✅,-,✅2023-09-18T00:00:00+0000,2023-09-18T23:59:59+0000, TUM_I, 228, 1440,✅,-,✅,✅,✅,-,✅The numbers in the columns “interferograms” and “ground_pressure” are the number of interferograms and the number of ground_pressure lines for the respective day.
Running the Retrieval in a Computing Cluster
You can of course run this pipeline on a computing cluster (e.g. SLURM-based).
Since the python cli.py retrieval start command terminates after starting the
pipeline in the background, any compute node would also terminate right away.
Therefore, you have to use the underlying call the the main.py script of the
retrieval module: python src/retrieval/main.py.
We use the following SLURM script on CoolMUC-4 at LRZ:
#!/bin/bash#SBATCH -J erp#SBATCH -o /dss/dsshome1/lxc01/ge69zeh2/Documents/em27-retrieval/em27-retrieval-pipeline/data/logs/%x.%j.%N.out#SBATCH -D /dss/dsshome1/lxc01/ge69zeh2/Documents/em27-retrieval/em27-retrieval-pipeline#SBATCH --clusters=cm4#SBATCH --partition=cm4_tiny#SBATCH --qos=cm4_tiny#SBATCH --time=06:00:00#SBATCH --nodes=1#SBATCH --cpus-per-task=112#SBATCH --export=NONE#SBATCH --get-user-env#SBATCH --mail-type=all#SBATCH --mail-user=moritz.makowski@tum.de
# setup environment: git is used to determine the commit hash of the currently# running pipeline, gfortan (gcc) is used to compile proffast and the ifg# corruption filtermodule load slurm_setupmodule load gitmodule load gcc/13.2.0
# activate virtual environment: we set up the virtual environment on the login# nodes, but you can also do that inside the compute nodessource .venv/bin/activate
# run retrievalpython src/retrieval/main.pySimpliy dispatch it with SLURM using:
sbatch cm4.sh # using your script nameAs of Pipeline version 1.6.2, the retrieval watcher also works, when the retrieval process is
running on a separate node using the --cluster-mode flag:
python cli.py retrieval watch --cluster-modeDon’t forget to increase the number of parallel processes of the pipeline with
config.retrieval.general.max_process_count 😉. We run 100 retrievals in parallel
on CoolMUC-4.